As One Health approaches become implemented more broadly, data analysis frequently is analyzed according to traditional siloed disciplines, which we believe negates the power gained by using a One Health approach to data collection. For instance, One Health research conducted traditional study designs from epidemiology or ecology may fail to support systems that require consideration of multiple species.


In this work we focus on designing systems for One Health research, from data design to data management to data analysis using joint inference.
Regarding study design, epidemiological methods assume that individuals or clusters are selected with a probability to population size. If population size and density between species (e.g. people and livestock) are not correlated or is unknown, then selecting one population will reduce or even negate the representativeness of the survey in another species. Ecological study designs often result in less representative data at the population level because the population size and density of many wild species is not known.
We developed a solution that integrates epidemiological and ecological study designs to improve representativeness of One Health studies of zoonotic diseases.
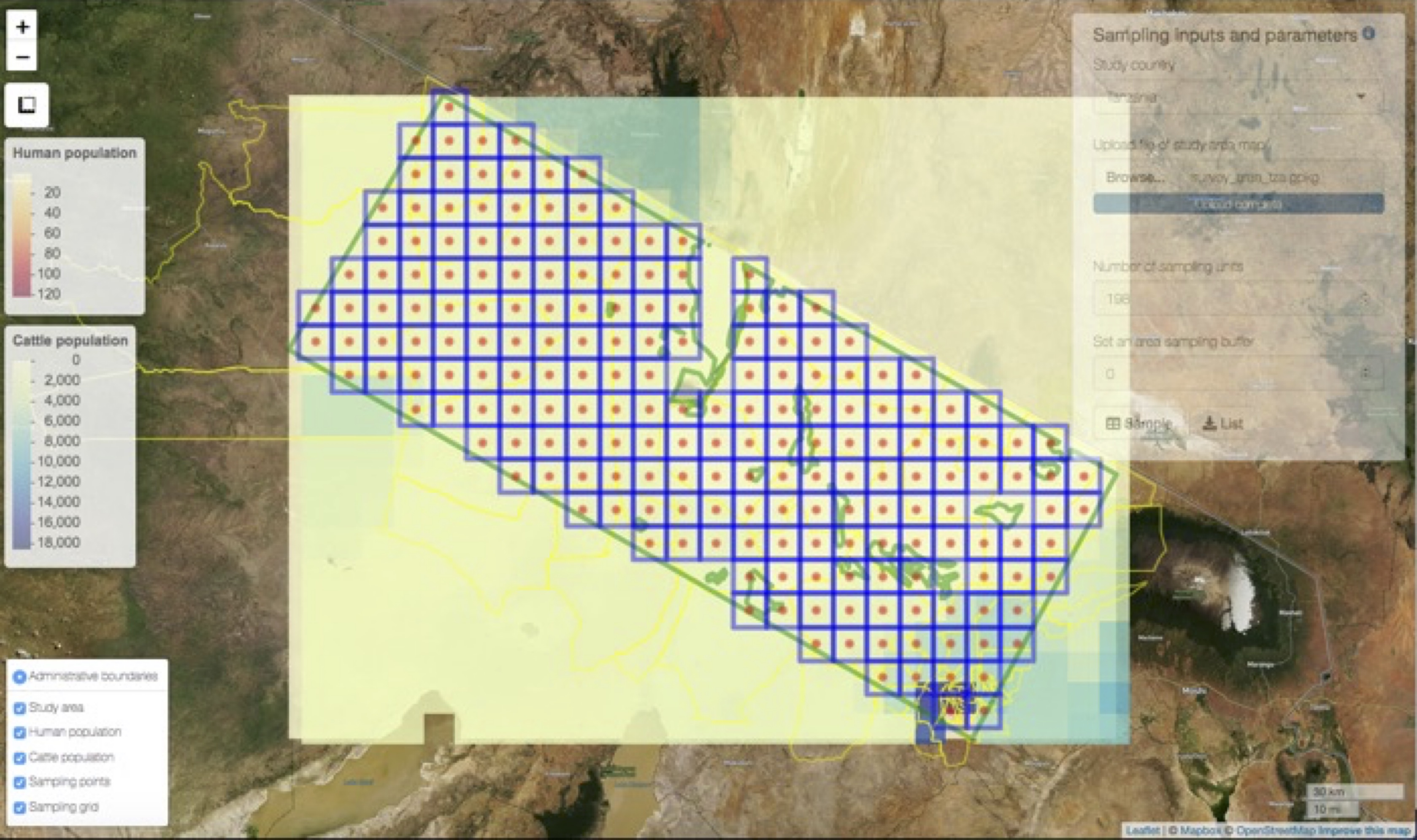 We used a systematic approach by applying a grid of geographic points over the study area to systematically sample the populations while providing for post-hoc weighting of each individual population by density.
We used a systematic approach by applying a grid of geographic points over the study area to systematically sample the populations while providing for post-hoc weighting of each individual population by density.
Data collection systems, quality assurance processes and integration vary across fields making it challenging to integrate One Health data within and between projects.
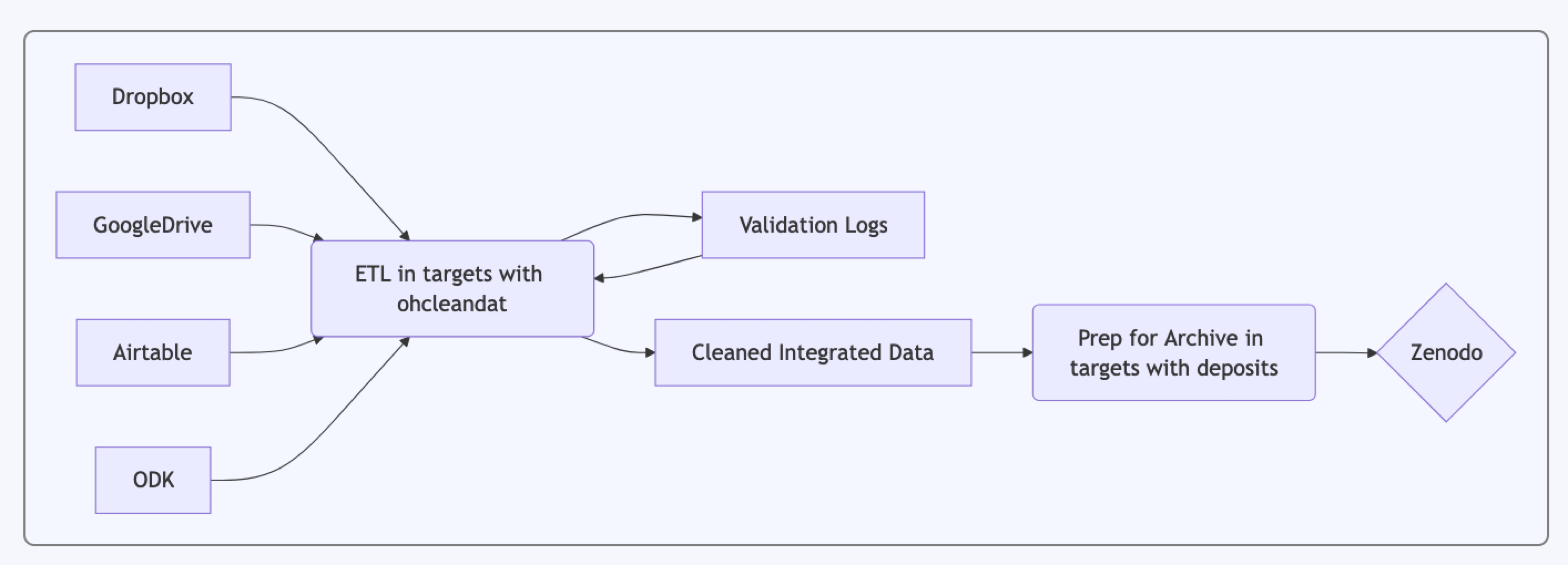
One Health data collected on different populations often use different methods and later need to be cleaned and integrated in a standardized way. To do this, we developed an R package, ohcleandat (One Health cleaning and data management system).
To support reproducibility, we used a targets workflow using the targets package (all in the R programming language) with the mechanics of ETL (Extract, Transform and Load) largely handled by the ohcleandat R package to manage the significant data repository, package. The underlying philosophy of the ETL process is the original data are only lightly modified (stripping white spaces, column names to snake case, etc.) while members of the project team make decisions about what data require cleaning and how they are to be cleaned. This is achieved by maintaining a validation log that keeps a record of all changes made and automates the process once the reviewer indicates a change must be made. Changes made in logs are applied to the data before they are integrated into various larger work packages. Those work packages are then deposited into Zenodo to create versioned single sources of truth with digital object identifiers.